What is different about the protein that is
destined for the rough endoplasmic reticulum?
[Note: This section describes work that led to a Nobel Prize in Medicine and Physiology to Dr. Gunter Blobel. For more information about Dr. Blobel's work and the pioneering discoveries, click: http://www.nobel.se/medicine/laureates/1999/ ]
The major difference is the fact that it has a hydrophobic signal sequence. This simplified cartoon shows that this is the first part of the protein produced. After the signal sequence is completed, protein synthesis is further inhibited. This is to allow the interaction of the signal sequence with a complex on the rough endoplasmic reticulum. In the above cartoon, note that the signal peptide is allowed to enter and essentially guide the protein into the lumen of the rough endoplasmic reticulum. Once the signal sequence is detected, protein synthesis resumes and the rest of the protein is inserted in the lumen. Note that a signal peptidase near the inner surface of the membrane works to cleave the signal sequence from the growing peptide.
The text reading for this discussion is Alberts et al, Molecular Biology of the Cell, third edition, Garland Publishing, 1994, pp 577-588 (Chapter 12) and pp 599-616.
 ]
]
The complex is actually more complicated than the above. The cartoon to the left shows a view of the signal sequence binding and interaction. Note that the signal sequence is recognized by a Recognition Particle, or SRP. This is then bound to a receptor. This complex guides the protein through a channel like region. It also consists of a docking site for the ribosome.

Another cartoon view of this process shows the signal receptor peptide (SRP) that associates with the large subunit of the ribosome that allows binding to the receptor on the rough endoplasmic reticulum.
After the protein is synthesized, the ribosome dissociates into large and small subunits and the SRP also looses its attachment to the receptor.
Current studies of ribosomal interactions with ER:
Andrea Neuhof, M.M. Rolls, B. Jungnickel, K-U Kalies and T A Rapoport, Binding of Signal Recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Molecular Biology of the Cell, 9: 103-115. 1997
- ØProteins destined for RER sorting make a signal sequence.
- ØAs signal sequence elongates, it is bound by the signal recognition particle (SRP54) (GTP dependent binding).
- ØSRP then binds to the SRP receptor (docking protein) on the ER membrane.
- ØAt the same time, ribosomes bind to the RER translocation channel formed by the Sec61p complex.
- ØThis Sec61p complex is a major component of a protein-conducting channel which also includes the “translocating chain-associating membrane protein” or TRAM. This conducts the protein into the RER sac.
Neuhof et al, 1997 have asked the following question: What stimulates the specificity of ribosomal docking?
- ØQuestion was asked because ribosomes dock to the Sec61p complex even without the signal sequence. Some clues:
- ØAs the signal sequence begins to appear (short) Øit can be removed from the ER membrane with high salt concentrations.
- ØAs the signal sequence elongates, Øribosome binding is stronger and ribosome complex becomes insensitive to proteases and high salt.
ØNeuhof's study hypothesized that the presence of the signal peptide was crucial for specific binding. Study Design:üAdded nontranslating ribosomes to compete with translating ribosomes in an in vitro system.
- üIf Signal receptor protein (SRP) was absent, üthe nontranslating ribosomes bound to the Sec61p receptor on ER membranes.
- üAlso, the nontranslating ribosomes competed with the translating ribosomes.
üAdded SRP to bind to the signal peptide.
- üThe translating ribosomes bound tightly and were not displaced.
- üThen, nontranslating ribosomes failed to compete for the Sec61p receptor sites.
Hence, SRP gives the translating ribosome a competitive edge once it starts translating the signal sequence.

You can see a better surface view in this cartoon. The cartoon is from your text. It shows the Ribosome sitting on the receptor next to the pore. The signal peptide is noted in red. The remainder of the code is read as the ribosome moves along the mRNA. The fluidity of the membrane allows the ribosome to be docked at its receptor site and also move along the mRNA. Each amino acid is added to the growing chain and the polypeptide gets longer.

This cartoon is also from your text. It shows the system without the ribosome. Here you get a better view of the pore through which the protein projects into the lumen and the signal sequence.
How are newly synthesized proteins inserted in the membrane?
Mechanism varies with the type of protein.
Type I: Signal sequence on amino terminus enters first and continues to elongate. Protein is threaded through the translocating channel (open area in rer membrane) until a hydrophobic stop sequence is reached. That hydrophobic stop sequence (seen as a hatched region in the protein) is then inserted in the membrane and forms the anchor for that protein. Signal is cleaved by protease inside the lumen. Type II: No cleavable signal sequence. These proteins have rather long hydrophobic regions that will be anchored in the membrane. Type II proteins are threaded into the lumen with the C terminus leading. Protein continues to be inserted until it reaches the hydrophobic stop signal sequence. Type III: Same as Type II, only the N terminus leads into the lumen.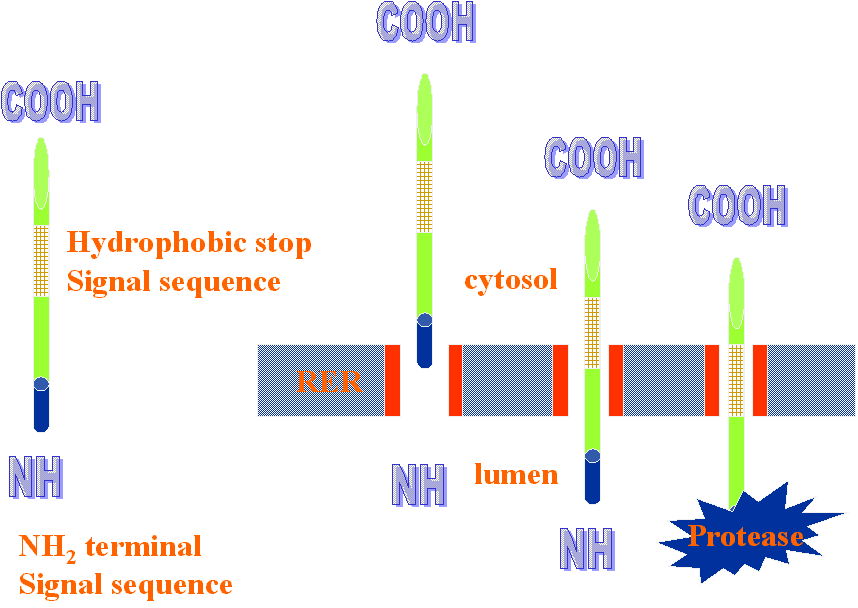

What regulates the orientation of Type II and III proteins?

Wahlberg, JM. And Spiess, M. Multiple determinants direct the orientation of signal anchor proteins: The topogenic role of the hydrophobic signal domain. J Cell Biology 137: 555-562.
Tested charged amino acids and length of hydrophobic signal sequence. The "positive inside rule" states that amino acid residues nearest the cytosolic side of the hydrophobic anchor sequence are more positive than those nearest the lumenal side. So, whichever end has the least positive charges near the signal anchor patch would go into the ER lumen. One can change the direction of translocation of a protein (reverse it) by mutating the protein and making more positively charged groups near the anchor patch of the other end. Below, the cartoon shows that this can be done to change a Type II protein (COOH end enters ER lumen) to a Type III (which has its amino terminal entering the lumen).
Washburn and Speiss (JCB 137: 555-562, 1997) also tested the length of the hydrophobic signal anchor sequence. The following cartoon shows that a longer hydrophobic anchor sequence (seen as the portion running through the membrane) promotes entry with the amino terminal leading into the lumen.

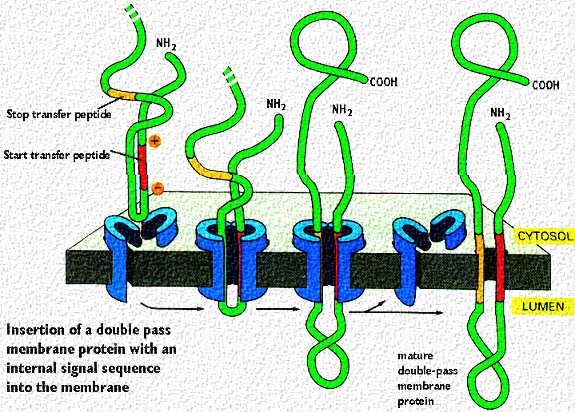
Proteins destined for insertion into membranes, such as ion channels or receptors have mRNA codes for start and stop sequences that allow multiple passes through the membrane. Signalling sequences (patches) can be formed as described in the above cartoon. It shows the insertion of a double pass transmembrane protein with the loop inside the rough endoplasmic reticulum. The red signal patch has the + charges near the cytosol and starts the insertion process. This continues until the hydrophobic stop signal patch is reached. That anchors the second membrane passage. Note, that both C and N terminal portions are in the cytosol Thus, if this protein is destined for the plasma membrane, that loop in the ER lumen will eventually project outside the cell
Membrane proteins that pass through the membrane multiple times (called "multipass transmembrane proteins) have multiple start and stop signals. They are aligned with the hydrophilic and hydrophobic portions of the lipid bilayer as described in the lecture on membranes. This is shown in the following cartoon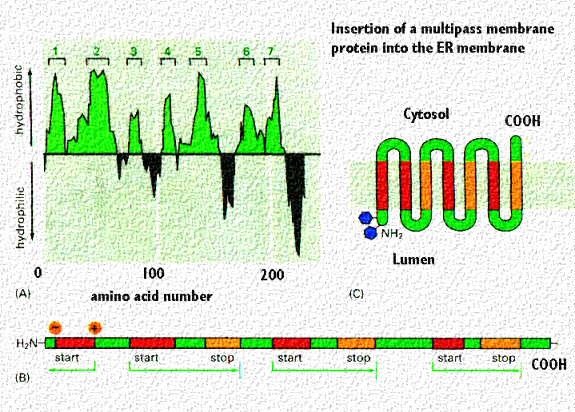
For more information, contact:
Gwen Childs, Ph.D.,FAAA
Professor and Chair
Department of Neurobiology and Developmental Sciences
University of Arkansas for Medical Sciences
Little Rock, AR 72205
For questions, contact this email address: